Unlike in other fields (i.e. genetics and cell biology), neuroscience does not have a standardized way to collect and share the wealth of existing data among researchers. The lack of a common format has made comparison across laboratories difficult and replication of specific experiments almost impossible, significantly slowing overall progress in the field.
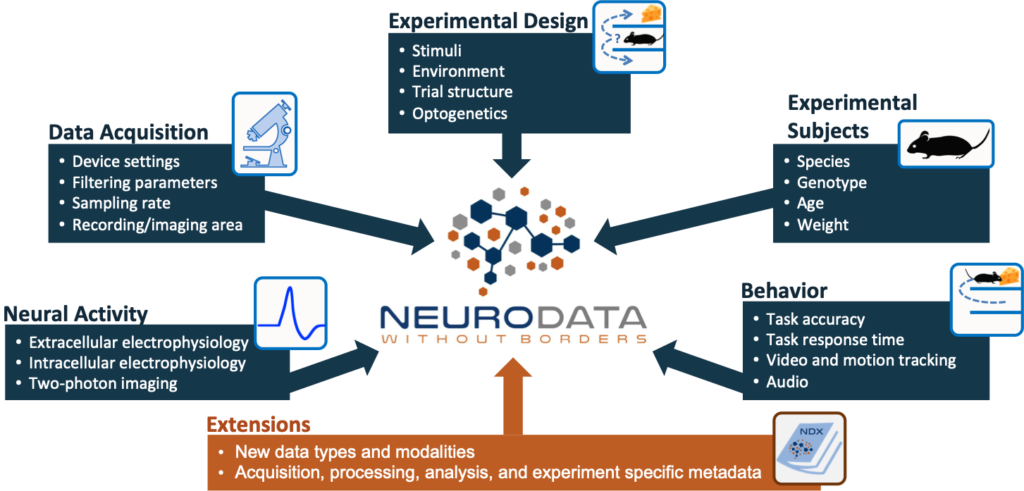
Neurodata Without Borders (NWB) is a data standard for neurophysiology, providing neuroscientists with a common standard to share, archive, use, and build common analysis tools for neurophysiology data. NWB is designed to store a variety of neurophysiology data, including data from intracellular and extracellular electrophysiology experiments, data from optical physiology experiments, and tracking and stimulus data. The project includes not only the NWB format, but also a broad range of software for data standardization and application programming interfaces (APIs) for reading and writing the data as well as high-value data sets that have been translated into the NWB data standard.
Neurodata Without Borders is intended to serve the broad neuroscience community and encourage the sharing of data by scientists worldwide. NWB 2.0 was released in February 2019. Please give it a try. Join our mailing list for updates or ask questions on our Slack channel.
To learn more about the approach taken to develop the NWB Format, please read our open access Neuron NeuroView article and bioRxiv preprint.
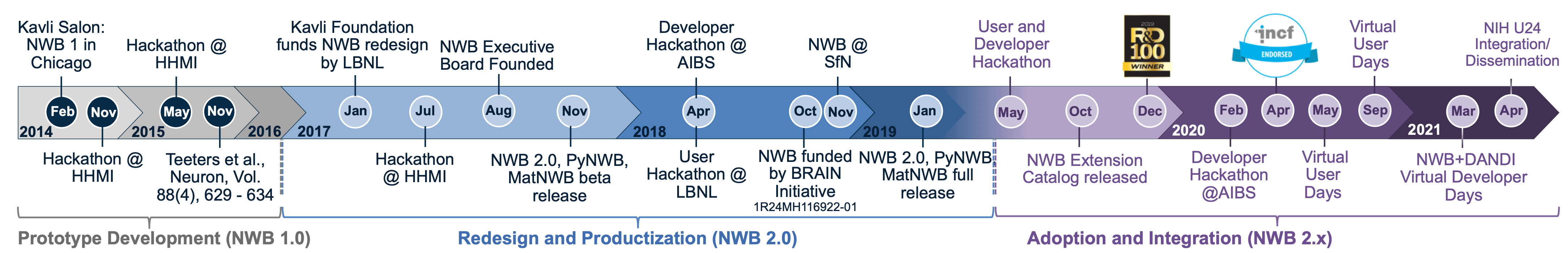
Launched in mid 2014, Neurodata Without Borders: Neurophysiology began as a pilot project to produce a unified data format for cellular-based neurophysiology data. NWB:N 1.0 was based on representative use case studies from four laboratories, and the project included a vetting phase for assessing whether other data models can also be used in the new common format. The initial development of NWB:N 1.0 was funded by industry and private foundations. Scientific partners include the Allen Institute for Brain Science (AIBS), the Svoboda Lab (Janelia), the Meister Lab (Caltech), the Buzsáki Lab (NYU), and Fritz Sommer/Jeff Teeters (UCB, maintainers of CRCNS.org). This one-year pilot project resulted in NWB:N 1.0.
Building on the success of NWB:N 1.0, the project entered a second phase, with additional leadership by Dr. Kris Bouchard, Dr. Oliver Rübel (Lawrence Berkeley National Laboratory, LBNL), Dr. Loren Frank (UCSF) and Dr. Edward Chang (UCSF). One main goal was to develop the next version of NWB:N and to enhance its adoption, through development of an advanced software architecture for NWB:N, a well-articulated data standards ecosystem, an open community software strategy, and advancements to the NWB:N data standard itself. NWB:N 2.0 was redeveloped by scientific software engineers/computer scientists at LBNL, Dr. Oliver Rübel and Mr. Andrew Tritt, in close collaboration with neuroscience laboratories and the broader NWB:N community. NWB:N 2.0 was released in February 2019 and new features continue to be added to the software ecosystem surrounding NWB:N 2.0.
The NWB:N project has also created a governance structure. An Executive Board (EB) decides the vision and roadmap for NWB:N and manages fundraising and outreach. The EB also works closely with a Technical Advisory Board (TAB), which makes technical decisions and maintains the software ecosystem.
We continue to see steady adoption of NWB:B by neuroscience labs, including the Allen Institute for Brain Science (AIBS), Bouchard lab (LBNL/UCB), Svoboda lab, Meister lab, Frank lab (UCSF), Chang lab (UCSF) and others. As labs adopt the format we also continue to see growing sets of data published in NWB. E.g, the AIBS has adopted the use of this format and to date has released numerous NWB:N files assaying mouse visual cortex.